Il programma di ricerca riguarda la identificazione di virus che infettano batteri, funghi e insetti (soprattutto vettori), per trovare nuovi metodi di lotta biologica a malattie delle piante e a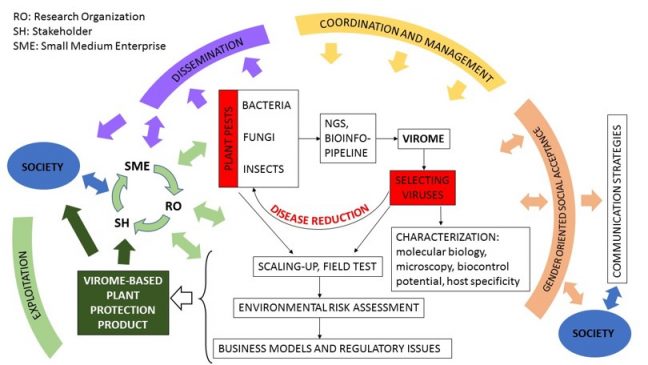 vettori di malattie delle piante. Inoltre, nello stesso progetto di ricerca si useranno i virus come piattaforma biotecnologica, attraverso piccole inserzioni genomiche di sequenze target, per sperimentare nuove biotecnologie per il contenimento delle malattie delle piante. Il programma prevede anche un work package sulla valutazione della fattibilità economica e uno sulla accettabilità sociale di nuove biotecnologie basate su virus da utilizzare in agricoltura.
vettori di malattie delle piante. Inoltre, nello stesso progetto di ricerca si useranno i virus come piattaforma biotecnologica, attraverso piccole inserzioni genomiche di sequenze target, per sperimentare nuove biotecnologie per il contenimento delle malattie delle piante. Il programma prevede anche un work package sulla valutazione della fattibilità economica e uno sulla accettabilità sociale di nuove biotecnologie basate su virus da utilizzare in agricoltura.
Coordinatore di Progetto: Massimo Turina
Partecipanti IPSP: Cristina Marzachì, Marina Ciuffo, Simona Abbà, Marta Vallino, Luciana Galetto, Marika Rossi, Andrea Delliri, Marco Chiapello
Sito web: www.viroplant.eu
Data inizio: 01/05/2018
Data fine: 31/10/2021
